- 2.1 The Search Page – Overview
- 2.2 Text Search Syntax
- 2.2.1 Booleans, Grouping, Phrase Searching
- 2.2.2 Wildcard Operators
- 2.2.3 Ontologies and Your Word Lists
- 2.2.3.1 Using Ontology Terms
- 2.2.3.2 Your Word Lists
- 2.2.3.3 Complete List of Ontologies
- 2.2.4 Constraining Searches to Specific Fields
- 2.2.4.1 Mixing Constrained Search and Other Features
- 2.2.4.2 Searching on Enumerated Fields
- 2.2.4.3 Searching on Date Fields
- 2.2.4.4 Searching on Numeric Fields
- 2.2.5 Proximity Search
- 2.2.6 Fuzzy Search
- 2.2.7 Boosting
- 2.3 Publication Number Search
- 2.4 Setting and Modifying Alerts
- 2.5 Tip Cards
- 2.6 Documents vs. Families
- 2.7 Saving Workfiles
- 2.8 Search Histories
- 2.9 Navigation Menu
2.1 The Search Page – Overview
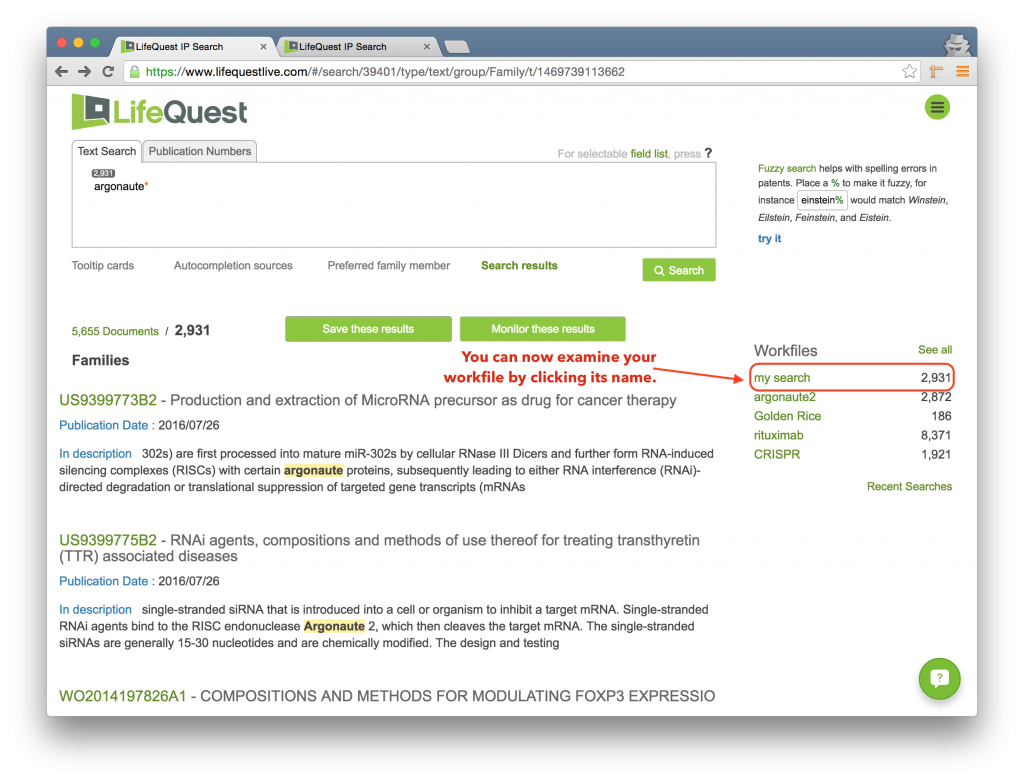
The first page that you are presented with in LifeQuest is the main search page. The purpose of this page is to allow you to construct a broad search so that you can save the results to a Workfile for more detailed review.
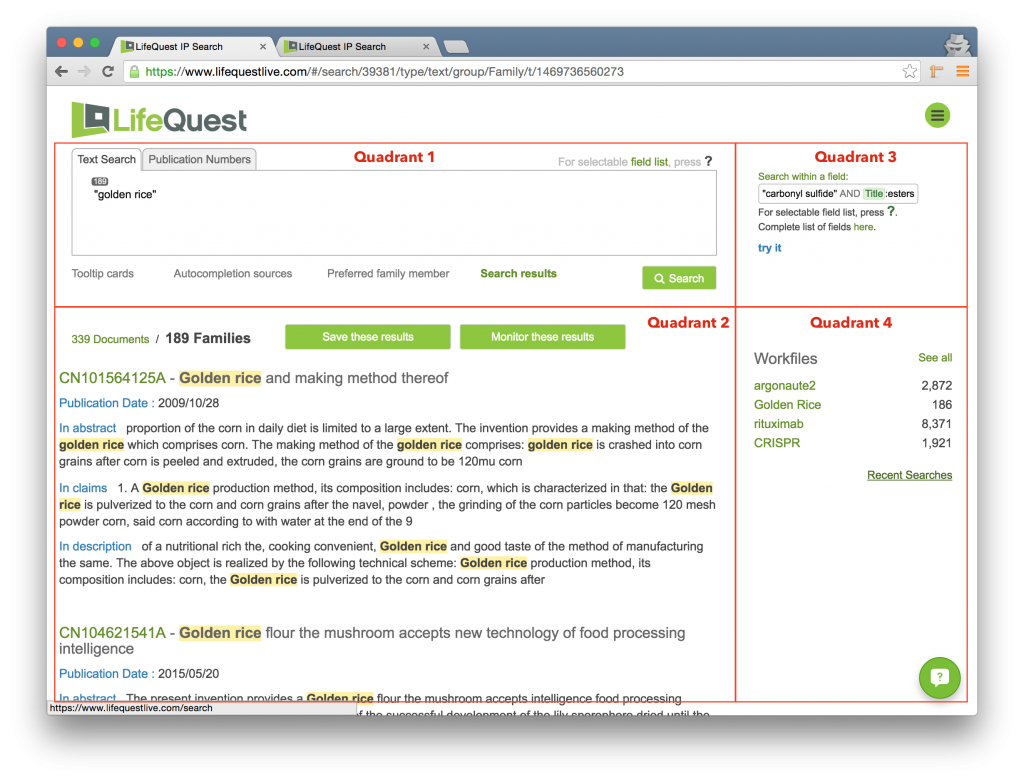
The page is divided into four primary quadrants, plus the LifeQuest logo (always clickable on every page and returns you to this search page) and the Navigation Menu, the green circle in the upper right.
| Quadrant | Contents |
|---|---|
| Quadrant 1 Upper Left |
The search box where you enter in your query. The syntax for queries is rich and documented here. Just underneath the search box are four selectable items: Tooltip cards, Autocompletion sources, Preferred family member, and Search results, each of which changes the content in the bottom left quadrant of the page. The Search results link won’t be clickable until you’ve actually run a search. |
| Quadrant 2 Bottom Left |
This quadrant will display tip cards, ontologies, preferred family member preferences, or search results depending on which of the four links just under the search box is selected. Two buttons entitled Save these results and Monitor these results allow you to create workfiles for further analysis and set email alerts to rerun the same search over time and be notified of changes. In the displayed image we are viewing the results of a simple phrase search for golden rice. |
| Quadrant 3 Upper Right |
In the top right quadrant LifeQuest provides a series of tips on the LifeQuest syntax. Click the try it link to copy the sample query into the search box. |
| Quadrant 4 Bottom Right |
The bottom right quadrant contains the list of workfiles, with links for interacting with individual workfiles, and managing workfiles, as well as the link to reach the Recent Searches page. The icon at the very bottom right requests real-time support via a chat window. |
2.2 Query Syntax

The LifeQuest Query Syntax is a modified Lucene-based grammar that should be familiar to a professional searcher. The Query Box itself is designed to carefully examine your input as you type. There are two very important indicators in the search box that should help you to build good searches. The first is the counter, a small number above your search. As you build your queries, this number will change to indicate the number of documents that will be returned when you execute your search. As you begin to using Grouping to build more complicated queries, the counter will compute the number of documents that will be returned by whichever group your cursor is currently in.
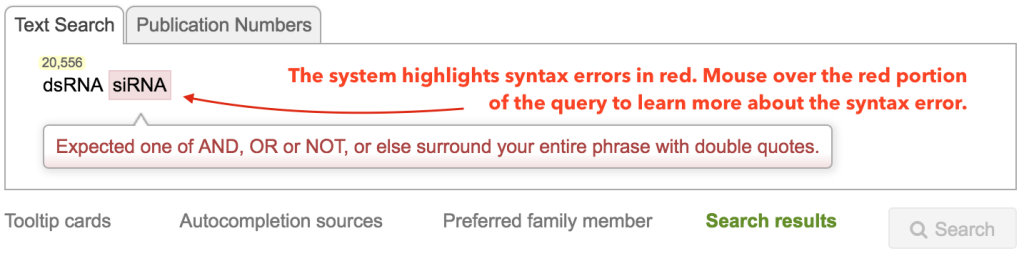
The second important indicator in the query box is the syntax error indicator. Whenever the system detects a syntax error, it will highlight the terms that are in error in red. By mousing over the red text you will get an explanation of the reason for the error. In the below example, the error is produced due to the ambiguity of the search – two terms next two each other must either be joined by a BOOLEAN such as AND, OR, or NOT, or searched as a phrase by using double quotes.
2.2.1 Booleans, Grouping, Phrase Searching
Boolean Operators
Each term in the query box must be joined together with a boolean operator. Three boolean operators are supported:
| Operator | Example | Definition |
|---|---|---|
| AND | dsRNA AND siRNA | Returns documents that have both of the terms |
| OR | cancer OR neoplasm | Returns documents that either or both of the terms. Documents with both of the terms will be ranked as more relevant than documents with just one of the terms. |
| NOT | NOT diagnostics | Returns documents that are specifically missing the term after the NOT. |
Note that there is no implicit Boolean operation occurring. So while you might type two terms next to each other such as:
you’ll notice that the siRNA is a syntax error. This is because LifeQuest wants you to be explicit – did you mean “dsRNA AND siRNA,” “dsRNA OR siRNA”, or simply the phrase “dsRNA siRNA,” with the two terms occurring one right after the other?
In some cases LifeQuest will attempt to infer implicit boolean operators. For instance, you may attempt to type two words next to each other and LifeQuest will insert an implicit AND automatically:
Note that in such cases the AND is a lighter grey and italicized, indicating that it was inserted implicitly. You won’t be able to edit it – you’ll have to put the cursor to the left of the rightmost term and then hit the backspace key. Best practice is to be explicit with all boolean operators.
Grouping
There is an implicit order of preference in Boolean:
- NOT
- AND
- OR
All of the NOTs are evaluated first, followed by all of the ANDs, followed by the ORs. In the below query:
we are asking for all documents that contain dsRNA and virus, unioned with all of the documents that contain viri but not siRNA. Using parenthesis to describe the order of operations, the above query would be expressed:
Indeed, the above two queries are equivalent.
You can overrule the implicit order of preference by using parenthesis. For instance, perhaps a more sensical query would have been:
Notice how we are using parentheses to group the terms whose implicit order of preference is not desirable, but we are still depending on the built-in order of preference for the NOT and AND terms in the above query.
Phrase Searching
If you want to search for a multi-word term (a “phrase”) such as “polycationic peptide,” place them inside of double quotes:
LifeQuest will return all documents that have these two words right next to each other. (For searches where you want these words to be close together but not necessarily right next to each other, see Proximity Searching.)
Of course, you can combine these phrase searches with other Boolean operators and with parenthetical grouping, for instance:
2.2.2 Wildcard Operators
There are two wildcard operators in LifeQuest, namely * and ?.
| Operator | Function | Commentary |
|---|---|---|
| * | Matches zero or more non-space characters. | When searching for vaccin*, the results will match any word starting with vaccin, e.g. vaccine, vaccines, vaccinated, vaccine2003, and others. |
| ? | Matches exactly one character. | When searching for H1N?, the results will match any 4 letter word starting with H1N, e.g. H1N1, H1N2, H1N7, and others. |
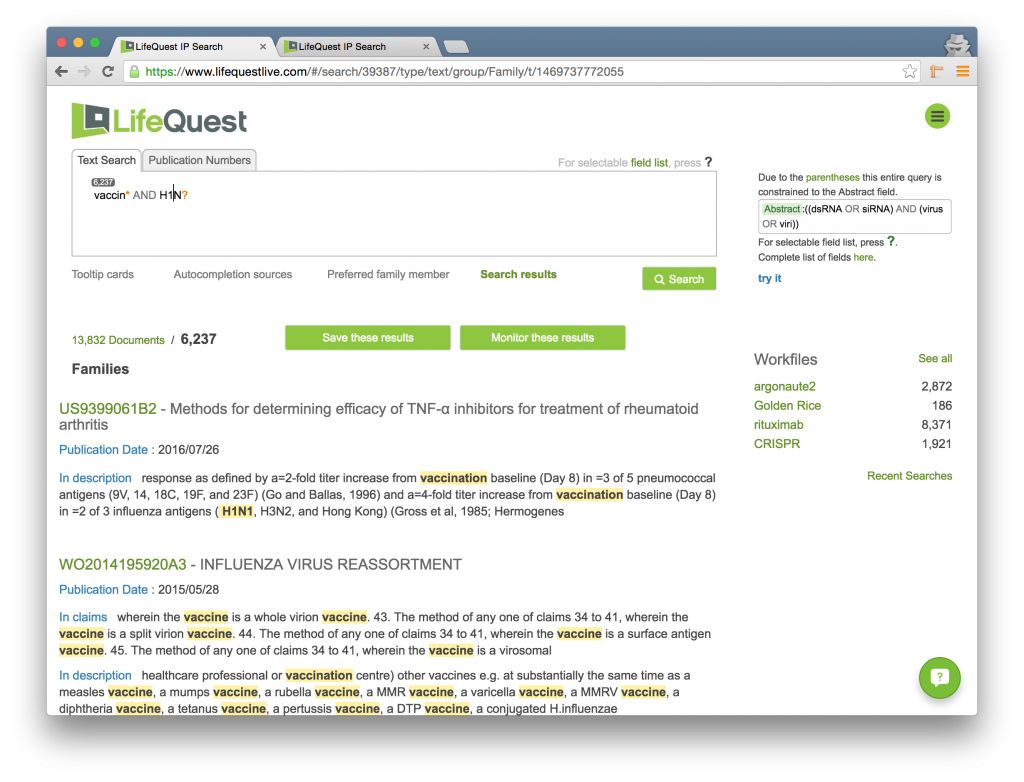
You can get arbitrarily complicated with wildcard use. As an example, they don’t have to occur at the end of a word. For instance, fol*ox? would hit to FOLFOX, FOLFOX4, and FOLFIRINOX. However, using either of the two wildcard symbols, ? or *, as the first character in your query greatly increases the complexity of the search and therefore the time it may take to return results.
Combining Wildcard Operators with Other LifeQuest Functions
Wildcard operators can be used in combination with other operations. For instance, you can combine wildcards with boosting:
The above query will match any word with the root antibod and boost the relevance of the document by a factor of 4 in comparison with matching the H1N1 term. See the documentation on Boosting for more information.
Another example of the use of wildcard operators in conjunction with other LifeQuest functions is its use in a field-constrained search. For instance, the below image shows how to find documents that start with any character and end with the word test but only in the Abstract. More on constraining searches to specific fields here.
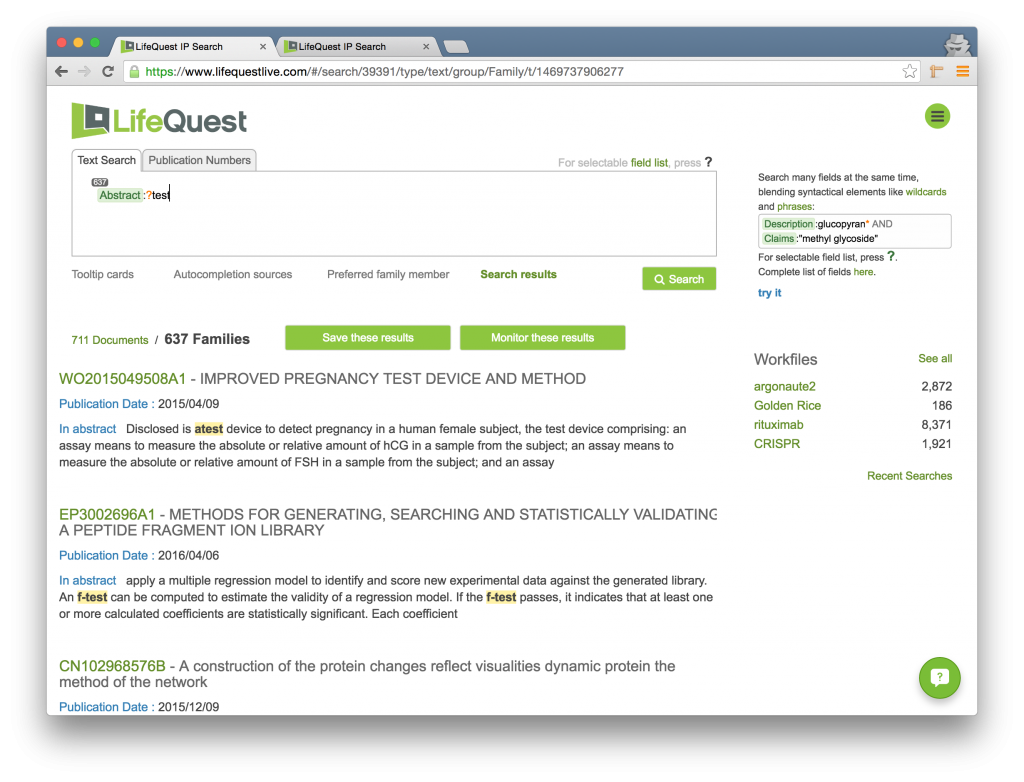
The ? is an Overloaded Character
Be advised that the ? character acts as a single character wildcard in the context described here. However, by hitting the ? key outside of a word you can bring up the field list to constrain your field search. Just keep typing and the field list will go away.
2.2.3 Ontologies and Your Word Lists
A key part of the LifeQuest system is its use of ontologies to allow you to search for concepts rather than just terms. The complete list of ontologies used by LifeQuest is available here.
2.2.3.1 Using Ontology Terms
LifeQuest automatically suggests ontology terms as you start typing. The default ontology selected is MeSH, or Medical Subject Headings, and as you type, LifeQuest suggests terms in that ontology. Watch this movie to see how this works in action.
As the user mouses over the ontology term s/he wants to search for, a window pops up indicating the different phrases that will be searched. While LifeQuest only displays the entry in the ontology, Alcoholism, it is in fact searching for every phrase enclosed in that term. By holding the mouse over the term one can see all of the phrases that will be searched.
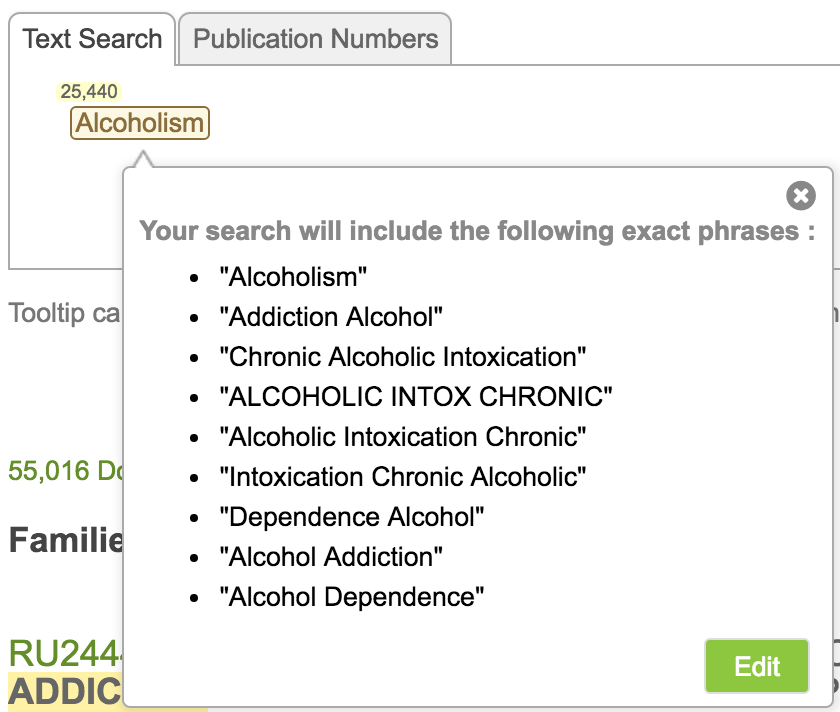
Indeed, searching this one ontology term is equivalent to the following search:
Multi-Word Ontology Terms
Let’s say you want to find the MeSH Term for dimethyl fumarate. If you type dimethyl and then hit the
The reason for this error is explained in the Booleans and Grouping section – LifeQuest doesn’t know whether you are joining these terms by a Boolean operator or whether you want to make this a multi-word string. The error forces you to be explicit.
To address this, start within double quotes! Note how this looks in the below picture. Because the multi-word string is within double quotes, the auto-completion is suggesting possible ontology terms you might mean, and no errors are showing.
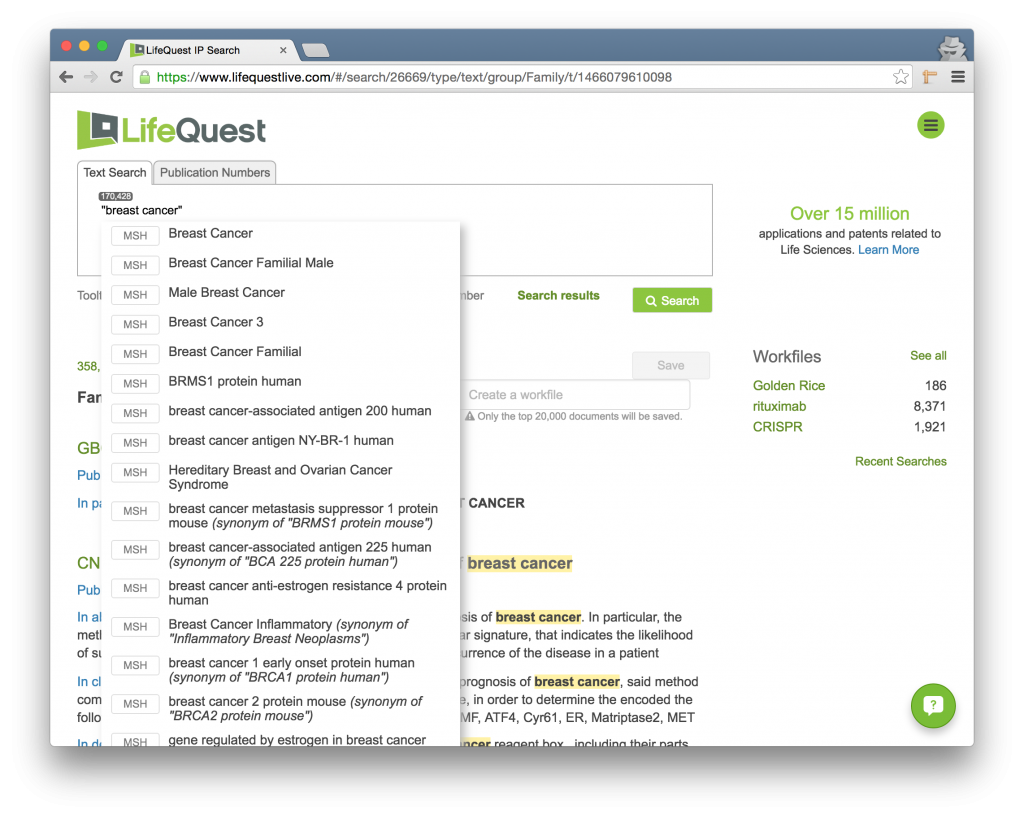
2.2.3.2 Your Word Lists
You can build your own ontology entries as well.
You can in fact edit ontology term descriptions to customize them for your liking. Once built, the system will try to auto-complete your queries using both the standard ontologies and your own word lists.
- Start typing in the search box until you find an ontology term that most closely matches the idea for which you are searching.
- Click on that term to insert the Ontology Term in the search box.
- Now, click on the Ontology Term to view its synonyms.
- Notice at the bottom of this list there is a button entitled Edit. Click this button.
- Edit the existing terms or add new terms, each phrase on its own line.
- Click save. Notice that the term now becomes blue, indicating it is no longer a stock ontology term, but rather a custom term owned by you.
The following video demonstrates this process.
2.2.3.3 Complete List of Ontologies
The complete list of ontologies is displayed in the bottom left hand quadrant of the search page when you click the link called “Autocompletion sources” which is just under the search box. You can toggle these on and off with the checkboxes to the left. Doing so will enable or disable the use of each respective ontology in suggesting terms for you as you type. You can in fact enable and disable these ontologies in the middle of creating a query, and you will notice how the terms suggested by LifeQuest change immediately as you toggle.
| Ontology | Abbreviation | Source | Description | Number of entries |
|---|---|---|---|---|
| Chemical Entities of Biological Interest | CHEBI | EMBL-EBI ChEBI | A dictionary of small molecular entities including any constitutionally or isotopically distinct atom, molecule, ion, ion pair, radical, radical ion, complex, conformer, etc. | 31,847 |
| Human Disease Ontology | DOID | University of Maryland’s Institute for Genome Sciences | A standardized ontology for human disease terms, phenotype characteristics and related medical vocabulary disease concepts. | 8,746 |
| Gene Ontology | GO | Gene Ontology Consortium | Controlled vocabularies that describe gene products in terms of their associated biological processes, cellular components and molecular functions in a species-independent manner. | 41,392 |
| Human Phenotype Ontology | HP | Human Phenotype Ontology | Phenotypic abnormalities encountered in human disease, e.g. atrial septal defect. | 10,593 |
| INOH Protein name/family name ontology | IMR | Multiple researchers in various Japanese organizations | A subsection of the now defunct database of signal transduction pathways known as INOH – this section protein names and their relationships to protein complexes in which they are known to be involved. | 9,217 |
| Protein Modification | MOD | HUPO Proteomics Standards Intiative | A vocabulary of protein modifications including those categorized by amino acid modifications, chemical processes, isobaric sets, and many others. | 2,001 |
| Medical Subject Headings | MSH | National Institutes of Health | The National Library of Medicine’s controlled vocabulary thesaurus. It consists of sets of terms naming descriptors in a hierarchical structure that permits searching at various levels of specificity. It is one of the most extensive ontologies in LifeQuest and has both very broad entries such as “Anatomy,” or “Mental Disorders,” as well as very specific entries such as “Ascorbic Acid.” | 343,688 |
| NCBI Organismal Classification | NCBI | National Institutes of Health | NCBI’s curated classification and nomenclature for all of the organisms in public sequence databases. | 821,808 |
| Plant Ontology | PO | Plant Ontology Consortium | A controlled vocabulary that describes plant anatomy and morphology and stages of development for all plants. | 1,356 |
| Protein Ontology | PR | Protein Ontology Consortium | A structured representation of protein forms and complexes. | 60,311 |
| Systematized Nomenclature of Medicine – Clinical Terms | SNOMEDCT | International Health Terminology Standards Development Organisation | SNOMED CT is a clinical vocabulary with global scope covering a wide range of clinical specialties, disciplines and requirements. | 332,278 |
| Plant Trait Ontology | TO | Gramene | A controlled vocabulary to describe each plant trait as a distinguishable feature, characteristic, quality or phenotypic feature of a developing or mature individual, for instance glutinous endosperm, disease resistance, plant height, photosensitivity, male sterility, etc. | 1,331 |
| Your word lists | USER | The growing list of word lists that you develop to manage your own search workflow. |
2.2.4 Constraining Searches to Specific Fields
Searches that you create will search the entire patent document – all of the title, abstract, description, claims, and bibliographic information. Often, however, you will want to constrain your search to specific fields within the system.
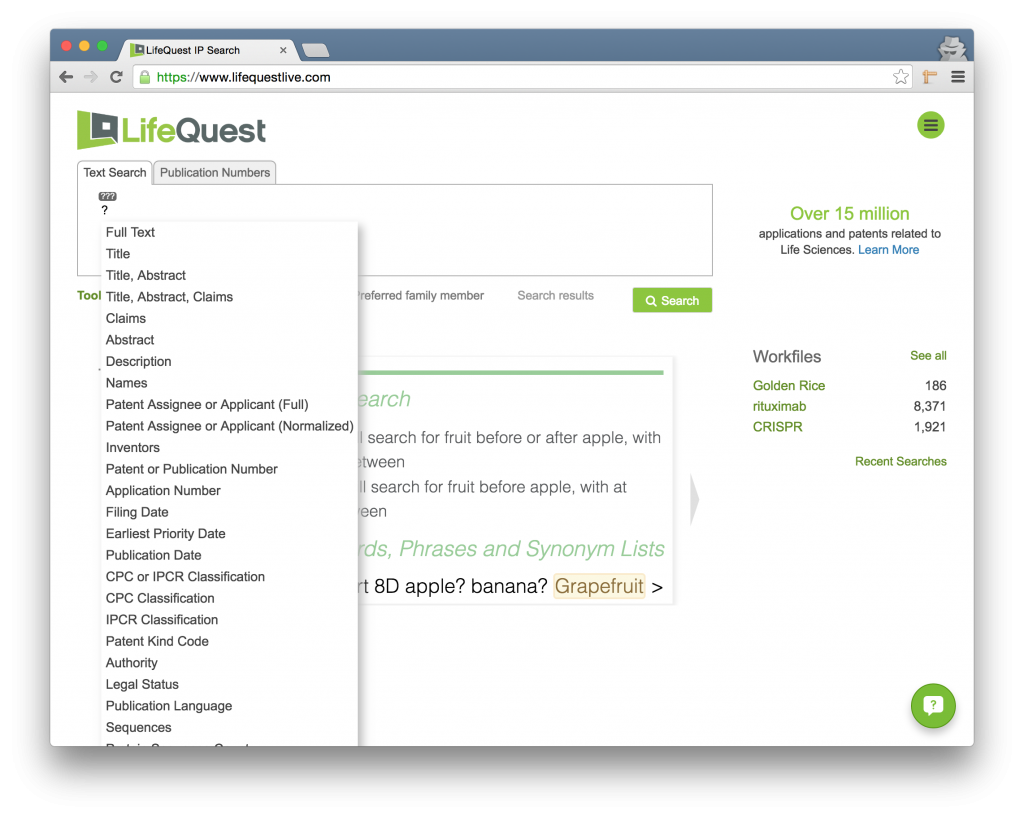
To constrain a search to a field, type the question mark ? key to pop open the field list. (Note that this operator is overloaded and also acts as a single-character wildcard if you use it within a word.) The next term you type will only be searched for in the field you chose.
For instance, the following search looks for the phrase “carbonyl sulfide” and the word mycobacterium in the title:
2.2.4.1 Mixing Constrained Search and Other Features
| Feature | Example and Commentary |
|---|---|
| Booleans | Claims:(apple OR banana) You can use an arbitary number of boolean operators in a constrained search, provided you enclose the entire query with parenthesis. |
| Grouping | Abstract:((dsRNA OR siRNA) AND (virus OR viri)) Grouping works exactly the same within a constrained field search as it does in an open search. Be sure to enclose the constrained query within an outermost parenthetical otherwise only the first term will be constrained to the specified field. |
| Phrase Searching | Title:“carbonyl sulfide” Simply place the phrase in double quotes. |
| Wildcard Operators | Description:glucopyran* AND Claims:GLP? This query will find documents that have words like glucopyranosyloxysalicylic and glucopyranoside mentioned in the Description, provided they ALSO have terms like GLP-1 or GLP-2 in the Claims. Note the combined use of multiple fields and the use of the Boolean operator AND to join. |
| Ontologies | Claims:succinyl group You can constrain ontology searches to fields as well. The above searches the claims for the ChEBI term “succinyl group” which in turn searches for many synonyms such as C4H4O2, butanedioyl, and others. |
| Proximity Search | Patent Assignee or Applicant: < Robert 3D Langer > This query will find patents whose Assignee and/or Applicant contain the words Robert and Langer within three words of each other, allowing for rearrangements of first and last name, middle initial, etc. |
| Fuzzy Search | Title, Abstract, Claims:metronidazole% Fuzzy search works perfectly well in a constrained field search. Simply use the fuzzy search operator, %. The above query would return documents that contain words like metronidazole, metronidazole, metronidazol, etc. |
| Boosting | Abstract:“recombinant heparin”^3 OR protease Abstract:(“recombinant heparin”^3 OR protease) You can boost the relevance of an entire constrained field search, or just certain elements of it, as shown above. In the first example, finding “recombinant heparin” in the Abstract is three times more relevant than finding the word “protease” anywhere in the document. In the second example, LifeQuest only searches the Abstract field for each of “recombinant heparin” and “protease,” with the relevance of “recombinant heparin” three times higher where it is found. |
| Combinations of the above | Full Text: (drought tolerance OR “drought recovery”^2) AND CPC Classification:“A01N63/00” AND Legal Status:grant% You can make arbitrary combinations using all of these rules. In the above example, we are searching for documents in CPC Class A01N63/00 that relate to the ontology term “drought tolerance” or the phrase “drought recovery” anywhere in the full text, with “drought recovery” being twice as relevant. The legal status of the documents should be “grant” but with the % fuzzy search operator we allow for minor misspellings of that word. |
2.2.4.2 Searching on Enumerated Fields
Certain LifeQuest fields have a constrained set of possible values – enumerated fields. They are:
| Field | Values and behavior |
|---|---|
| Authority | After you select the Authority: tag, begin typing an Authority Name and a pick list will pop up that helps you complete the search. |
| Language | Similar to Authority, simply type the language after the Language: tag and the system will provide completions for you as you type. |
| Sequences | Whether the patent is found in the GenomeQuest sequence database or not. This field is constrained to either a yes or a no. The system will provide a pick list for you after you type the Sequences: tag. |
| Legal Status | The legal status of the document, one of Grant or Application. The system will provide a pick list for you after you type the Legal Status: tag. |
2.2.4.3 Searching on Date Fields
Some of the fields in LifeQuest contain dates, for instance:
- Filing Date
- Priority Date
- Publication Date
Date formats are YYYYMMDD. When you enter a date, LifeQuest automatically redraws it to indicate it understands you have entered a date. So if you type 20090214, LifeQuest will display it as: 2009•02•14.
There are two ways to search date fields. You can either seek an exact match, or you can search for a range.
Exact Date Matching
To find all documents with the Publication Date of January 1, 2010, constrain your search to that field and enter the date. You don’t have to type the bullets, just type 20100101 and LifeQuest does the rest:
Date Ranges
The other facility to search date fields is to enter a range of dates. To do this, constrain your search to the field in question and then use brackets [] to enter a range:
You can use ranges to construct any form of a date query, as described in the below table. In each case we assume we are searching on Publication Date, but you can constrain the search to any field you desire.
| Use Case | Example |
|---|---|
| All docs with pub date of July 4, 1976. | Publication Date:1976•07•04 |
| All docs after July 4, 1976, inclusive. | Publication Date: [ 1976•07•04 TO now ] |
| All docs after July 4, 1976, exclusive. | Publication Date: [ 1976•07•05 TO now ] |
| All docs with pub date before July 4, 1976, inclusive. | Publication Date: [ * TO 1976•07•04 ] |
| All docs with pub date before July 4, 1976, exclusive. | Publication Date: [ * TO 1976•07•03 ] |
| All docs with pub date between January 1, 1971 and July 4, 1976, inclusive. | Publication Date: [ 1971•01•01 TO 1976•07•04 ] |
2.2.4.4 Searching on Numeric Fields
Some of the fields in LifeQuest are numeric, for instance:
- Nucleotide Sequence Count
- Protein Sequence Count
Searching numbers are exactly like searching dates – you can either seek an exact match, or you can search for a range.
Exact Numeric Matching
To find all documents with no nucleotide sequences, constrain your search to that field and enter the number 0.
Numeric Ranges
The other facility to search numeric fields is a range. To do this, constrain your search to the field in question and then use brackets [] to enter a range:
You can use ranges to construct any form of a numeric query, as described in the below table. In each case we assume we are searching on Nucleotide Sequence Count, but you can constrain the search to any field you desire.
| Use Case | Example |
|---|---|
| All docs with exactly 100 nucleotide sequences. | Nucleotide Sequence Count: 100 |
| All docs with 100 or more nucleotide sequences, inclusive. | Nucleotide Sequence Count: [ 100 TO * ] |
| All docs with 100 or more nucleotide sequences, exclusive. | Nucleotide Sequence Count: [ 101 TO * ] |
| All docs with less than or equal to 100 nucleotide sequences. | Nucleotide Sequence Count: [ * TO 100 ] |
| All docs with less than 100 nucleotide sequences. | Nucleotide Sequence Count: [ * TO 99 ] |
| All docs with 50 to 100 nucleotide sequences, inclusive. | Nucleotide Sequence Count: [ 50 TO 100 ] |
2.2.5 Proximity Search
Often you may wish to search for the occurrence of a set of words that are in close proximity, but not necessarily right next to each other. LifeQuest allows this with either the ordered or unordered proximity operators, kW and kD, respectively, where k is the number of terms you wish to allow between the two search terms:
The above query finds documents where no more than 10 words occur between the occurrence of the terms diverticulitis and constipation, irrespective of which term is found first.
Similarly, the above query does the same however the term diverticulitis must precede the term constipation.
One way to think about the proximity operator is that exact matches have a proximity of zero, for instance, < h1n1 0W vaccine > is precisely equivalent to “h1n1 vaccine”.
In the same manner, word transpositions are at proximity 1, e.g., < vaccine 1D h1n1 > would find “h1n1 vaccine”.
Amplified AND
A query such as < foo 10000000D bar > is an interesting alternative to foo AND bar. Both queries will return essentially identical sets of documents, however the proximity query will assign a higher score to documents for which the terms foo and bar are closer together. In this way, the order of the documents returned will be different. The proximity query will be more expensive computationally than the straight boolean query, and will take longer to run, but may provide more relevant results at the front of the search.
Wildcards in proximity search
Proximity search supports wildcards directly: < brca? 5D diagnos* > returns documents containing either BRCA1 or BRCA2 within five words of any term starting with the letters diagnos, for instance, diagnose, diagnostic(s), diagnosing, etc.
Ontologies and Proximity Search
Instead of individual terms, you can use ontology entries in proximity search. All of the synonyms for that ontology term will then be used to run the search. For instance:
This will find any documents where any of the synonyms of MECA-79 antigen are found within 10 terms of any term beginning with the word LECAM
Phrases and Proximity Search
Phrases can also be employed in LifeQuest proximity search:
This returns any documents where the exact phrase leukocyte extravasation is found within 10 terms of the exact phrase cellular adhesion.
Constraining Proximity Search to Specific Fields
Proximity searching can be constrained to a specific field in the same manner as any other kind of LifeQuest search.
Multiple Term Proximity Search
You can place multiple terms on either side of proximity search operator. For instance:
This query will find documents where either the term endothelium or the term epithelium is found within 20 terms of either the term COPD or the phrase chronic obstructive pulmonary disease. Note that it functions interchangeably with terms or phrases.
Proximity Search – Putting it all together
All of the ideas described here can be combined to make arbitrarily complex queries.
2.2.6 Fuzzy Search
LifeQuest supports the idea of a fuzzy search, which allows for matches to words that are similar to but not exactly the same as the word you are searching. Fuzzy searching uses the Levenshtein Edit Distance concept to return hits that are equal to or have an edit distance of 1 from your search term.
The operator for fuzzy search is the %. It can only be applied to a single term. It can not be used for phrase searching. It also can not be used inside of a User Word List. However, it can be used in a query that is constrained to a specific field. For instance, consider the following search, which is constrained to the Inventor field.
This will return documents where the word einstein appears, but also where words that can be generated by making a one character insertion, deletion, or substitution in the word einstein. For instance:
| Word | Edits |
|---|---|
| Winstein | Substituted a W for the first E |
| Eilstein | Substituted an L for the N |
| Feinstein | Inserted an F at the beginning |
| Eistein | Deleted the N |
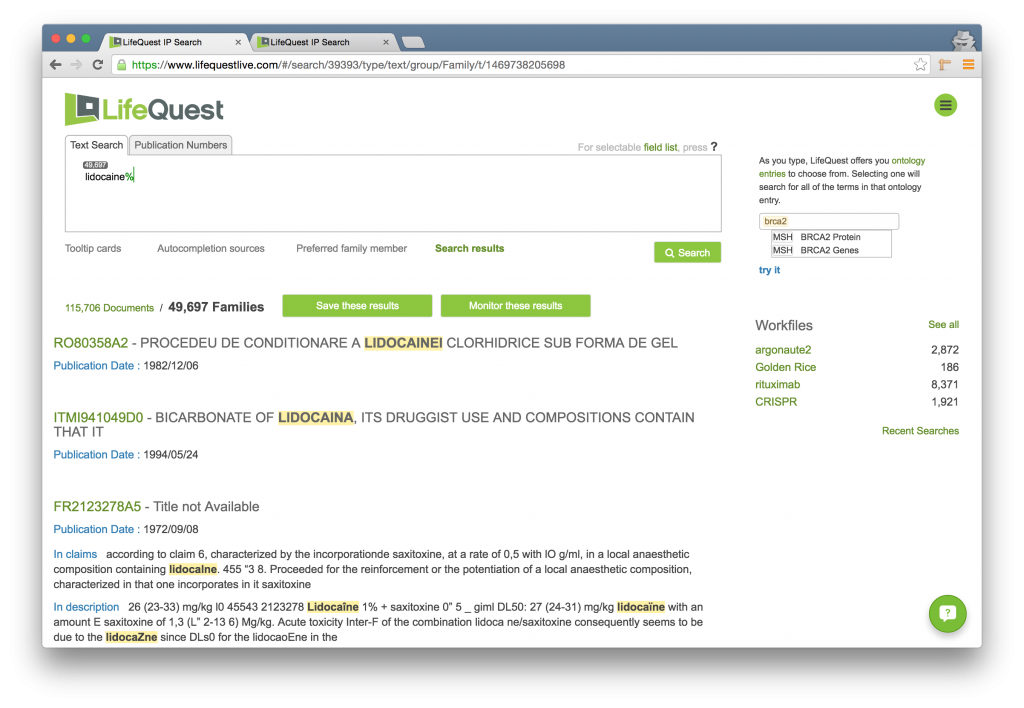
This technique is very useful when searching for terms where you suspect there may be minor misspellings due to automated translation into English. It is often used as a double-check to identify documents that might be slightly incorrect, as well as to build your own word lists for such misspellings. For instance, suppose you are seeking documents pertaining to LIDOCAINE. The following search shows all of the documents that refer to misspelled or foreign language spellings of the word, omitting those spellings that are correct:
Edit Distance Parameterization
LifeQuest supports fuzzy searches for terms with Edit Distances of either 1 or 2. The default is 1. To turn it to 2, add the number 2 after the operator:
would match to acidosis, acidosic, acidosises, acidolysis, etc.
Values greater than 2 are not currently supported – they will automatically be adjusted to just 2 edits.
2.2.7 Boosting
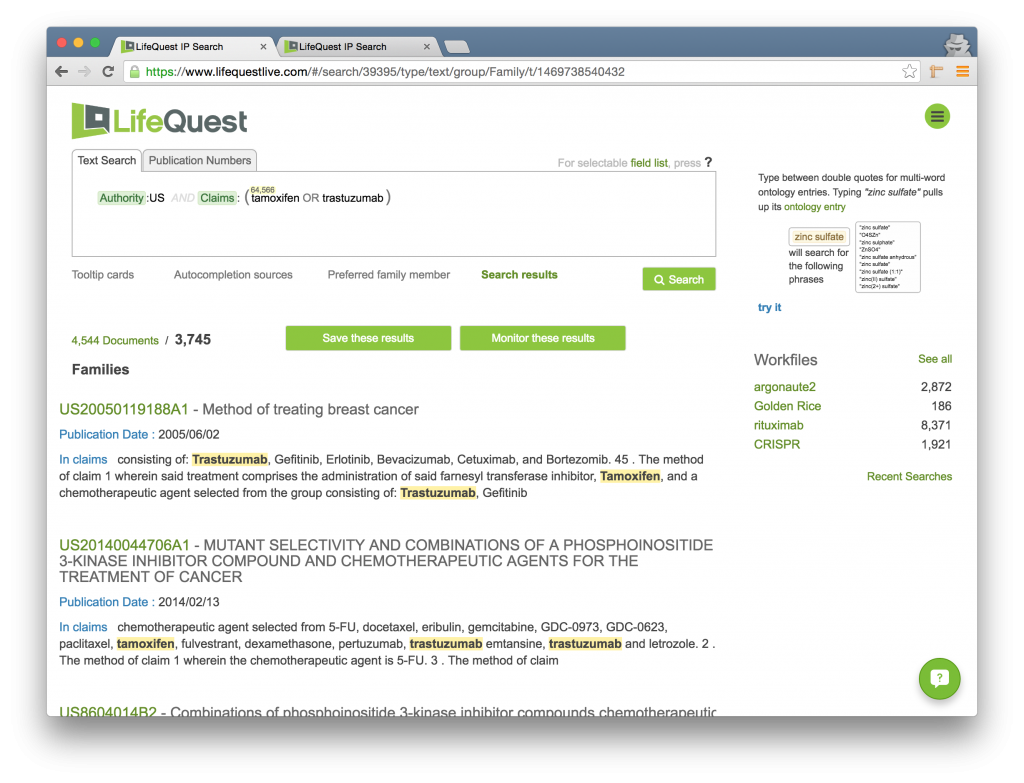
Boosting is a way to increase the relevance of certain documents in your search without changing the total composition of the search result. LifeQuest enables you to change the order in which documents are returned by changing the relevance of specific terms in your query. This is done by following a term, phrase, or Boolean group by the boosting operator followed by a number, e.g., tamoxifen OR trastuzumab^2, to mean that TRASTUZUMAB is twice as important in relevance ranking as compared to TAMOXIFEN. Consider the following query:
It asks for all US documents where either TAMOXIFEN or TRASTUZUMAB (or both, of course) are found in the Claims section of the document. When we run this query, we get 3,745 families returned. Note the top result is US20050119188A1 which contains both terms in the claims.
Now, we will boost the importance of the term TAMOXIFEN, making it ten times more relevant than TRASTUZUMAB, by using the Boost operator followed by the number 10:
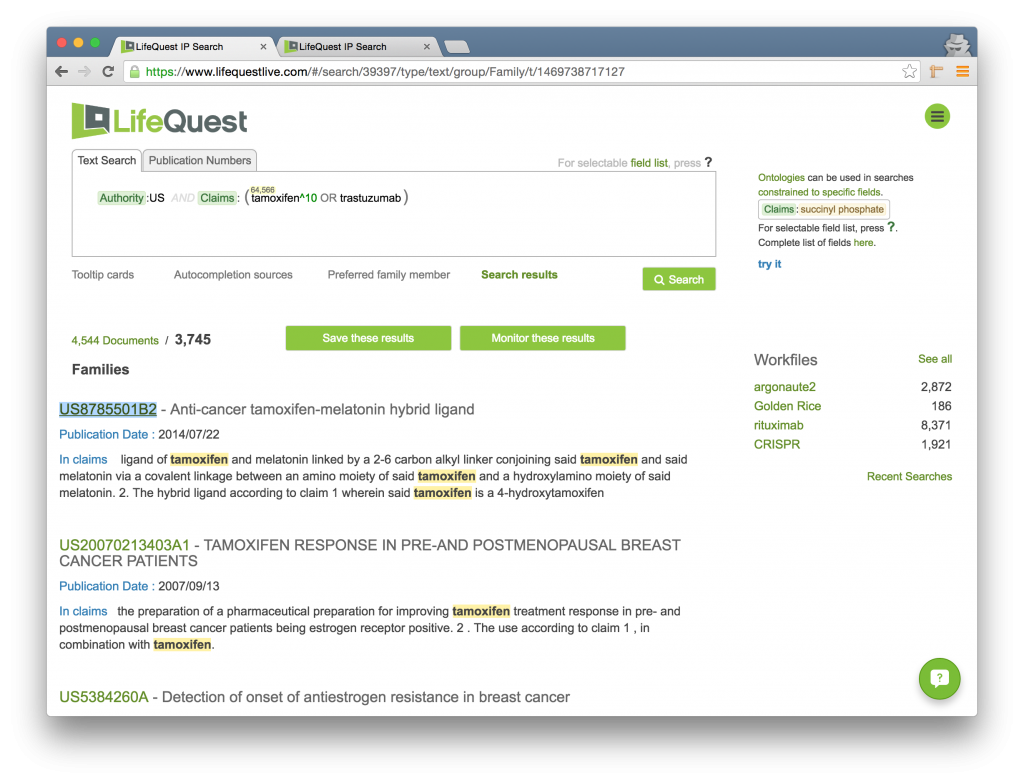
Notice that the number of families returned does not change. It is still 3,745. However, the first result in the set is no longer US20050119188A1, but rather US8785501B2, which only has TAMOXIFEN in the claims.
It is critical to understand that boosting does not change the ultimate composition of your result set – the exact same documents are returned whether you boost or don’t boost. The only difference is the order of the documents.
You can boost any term, phrase, or Boolean group. You can also boost a term or a group that is part of a constrained search, for instance, Authority:(US^2 OR EP). This operator can not be used inside of a User Word List.
2.3 Publication Number Search
2.4 Setting and Modifying Alerts
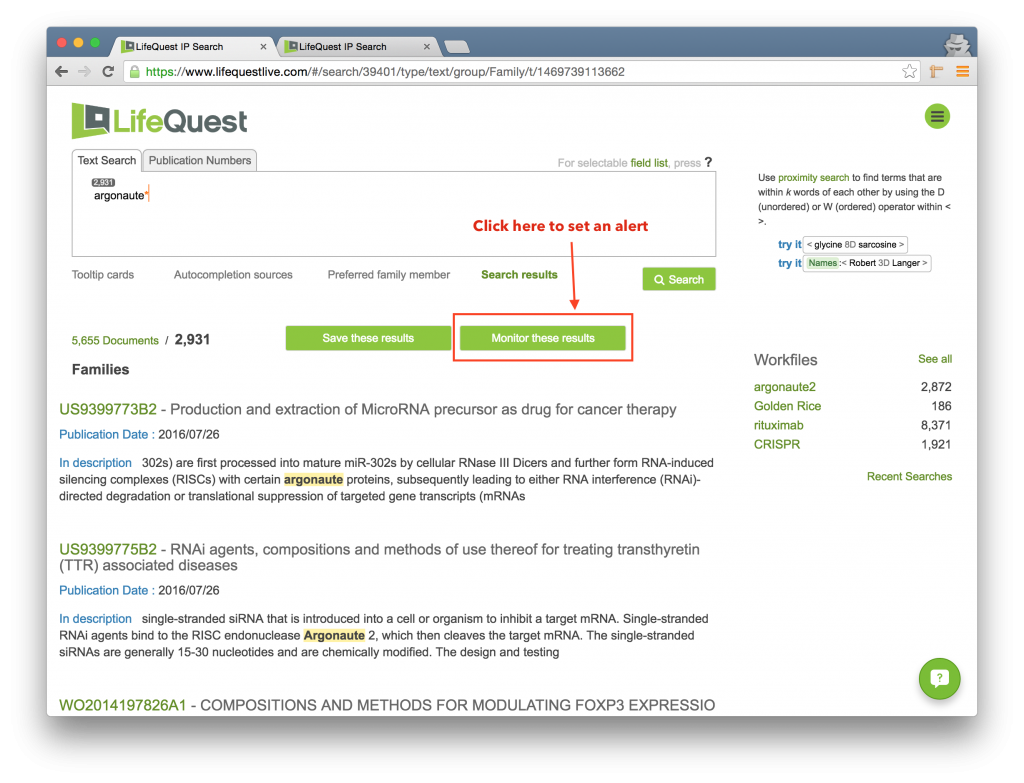
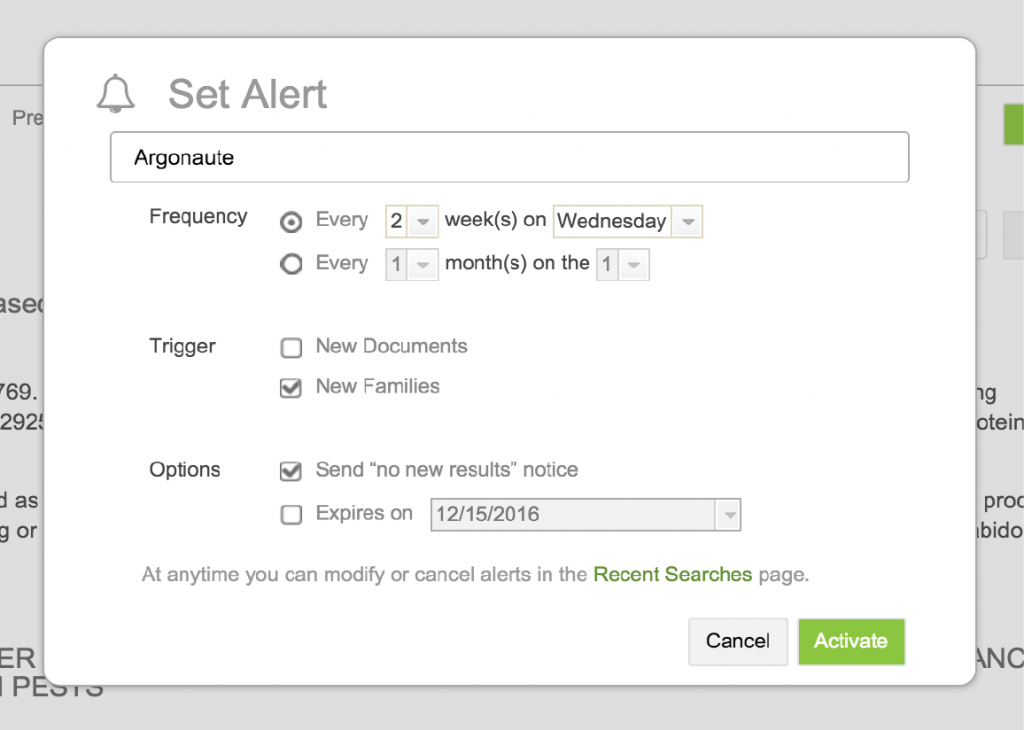
LifeQuest can automatically re-run any search you’ve submitted on a regular basis. If the search returns new results on any re-run, LifeQuest will email you with the updates. To set an alert, first run a search, and then click the Set Alert link just over the search result.
A dialog box will appear allowing you to configure the alert. You will be able to:
- Set the frequency – every i weeks on a particular day of the week, or every j months on a particular day of the month
- Set your notification preferences for new results: learn about any new documents, any new families, both, or the absence of any new results
- Set an optional expiry for the alert
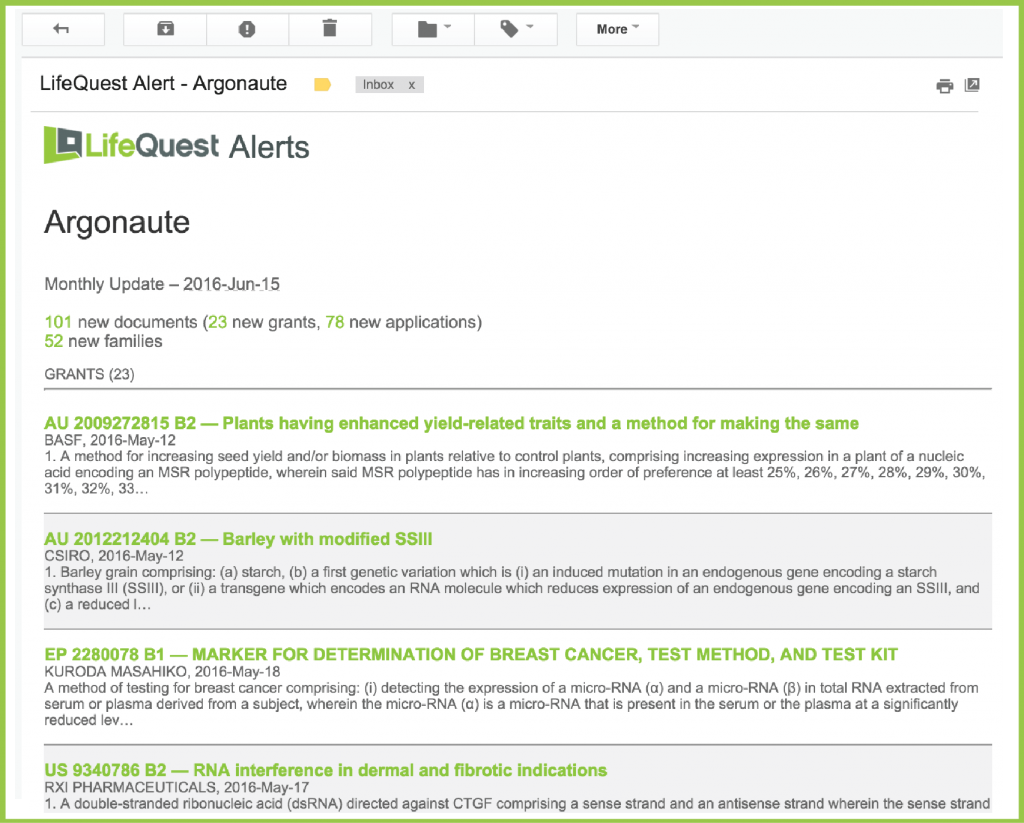
A typical email for an alert will contain first the list of new documents, if any, and then the list of new families:
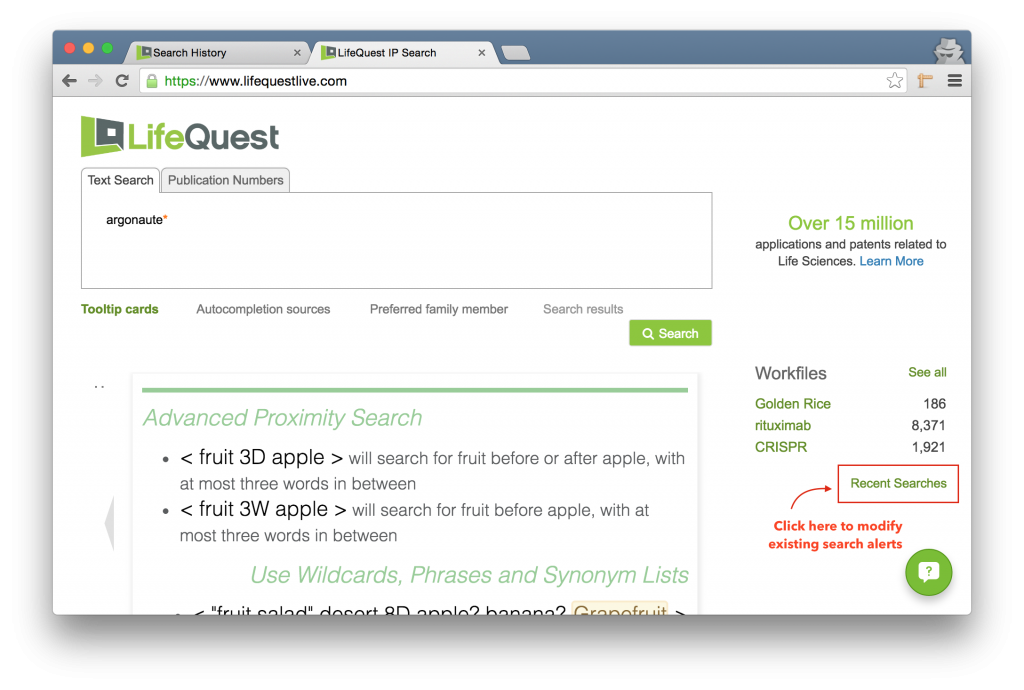
To modify an alert you have set up previously, click the Recent Searches link underneath the list of workfiles.
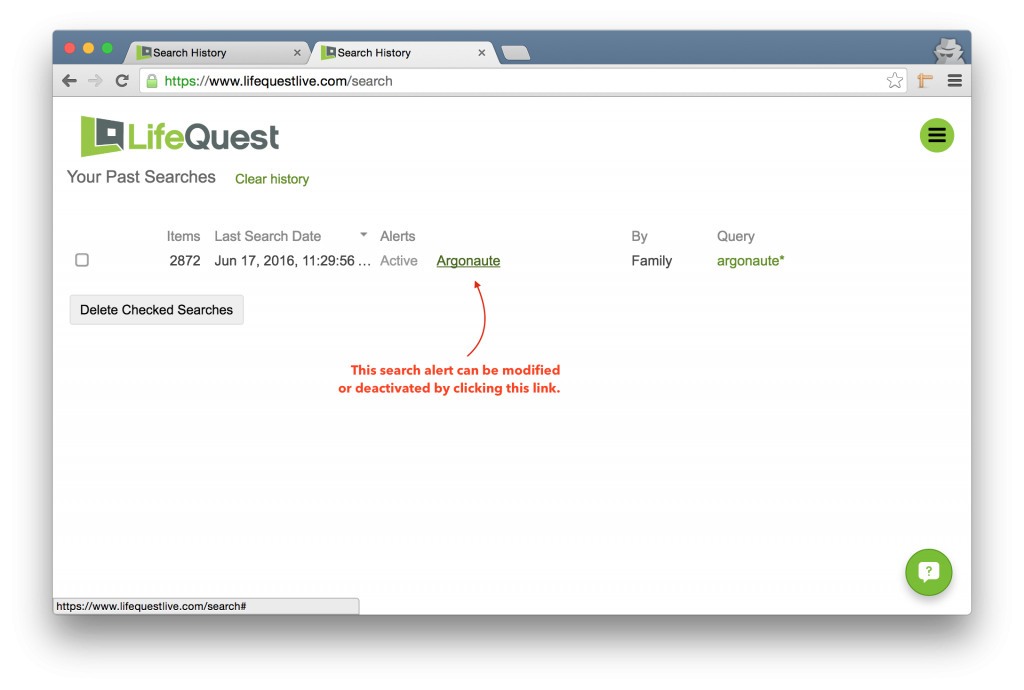
Then click the name of the alert to modify or deactivate it.
2.5 Tip Cards
Just underneath the search box you will find the Tip Card Area. This is series of small cards that provides a reminder of the LifeQuest search syntax.
Just to the left and right of the Tip Card Area are two small arrows. Clicking these will allow you to page through the different tips so that you can quickly find a syntactic example relating to your query.
Note that after you run a query, the Tip Card Area will be replaced by the Search Results. In order to bring the tip cards back, click the Tooltip Card link just underneath the search box.
2.6 Documents vs. Families
2.6.1 Toggle between Documents and Families
You have the ability to toggle search results to list either every document that matches your search sorted by relevance, or to list the single best representative per family that matches your search. This distinction comes into play when creating Workfiles, particularly those that will be merged with other workfiles, because those operations are performed solely at the Document rather than the Family level.
Note that the below search finds 11,476 families and 16,384 documents within those families. Presently we are viewing the families but by clicking the 16,384 Documents link the search would return all of the individual documents rather than the families.
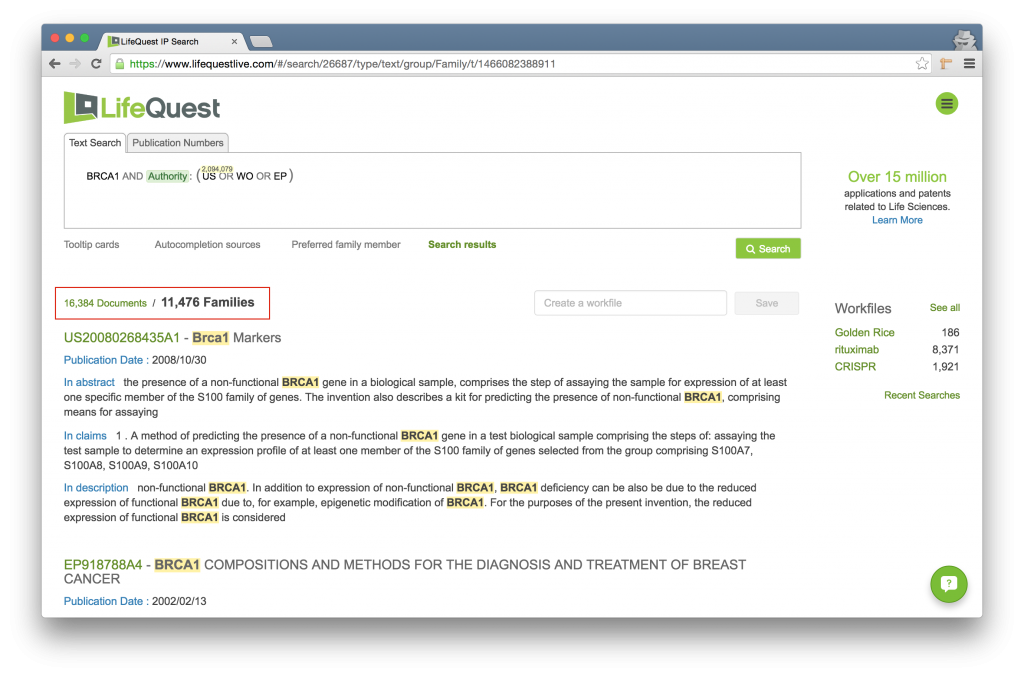
Note that this toggle uses the Extended Family definition employed by LifeQuest.
2.6.2 Preferred Family Member
When in Family Mode, that is, viewing the search results by family, which document of the family is displayed and saved in a Workfile?
This is a configurable option. By clicking the Preferred family member link under the search box, you can configure the right representative for each family.
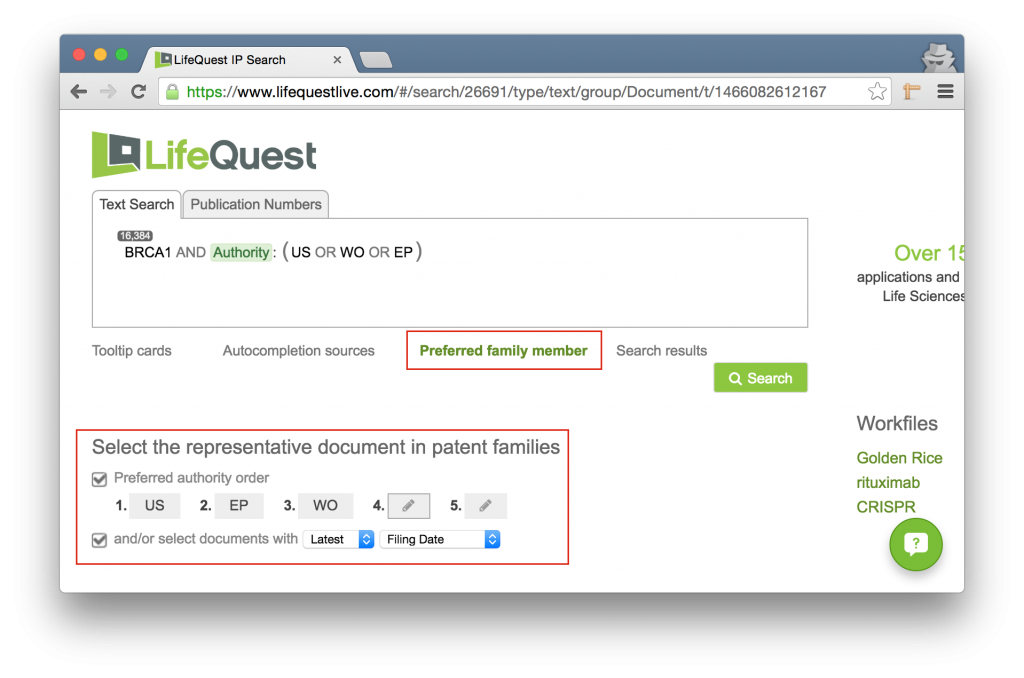
LifeQuest will select the representative document for a family based on either a preferred authority or by a set of dates. For instance, in the above example, if there is a US document in the family it will first choose that. If not, it will look for an EP document. If none, a WO. Finally, it will revert to the document with the latest filing date. In the event of more than one document from the highest preferred authority, it will again revert to the preferences around the date, in this case, latest filing date.
2.7 Saving Workfiles
You can save search results as a workfile to enable deeper analysis. To do this, click the Save these results button in the middle of the application.
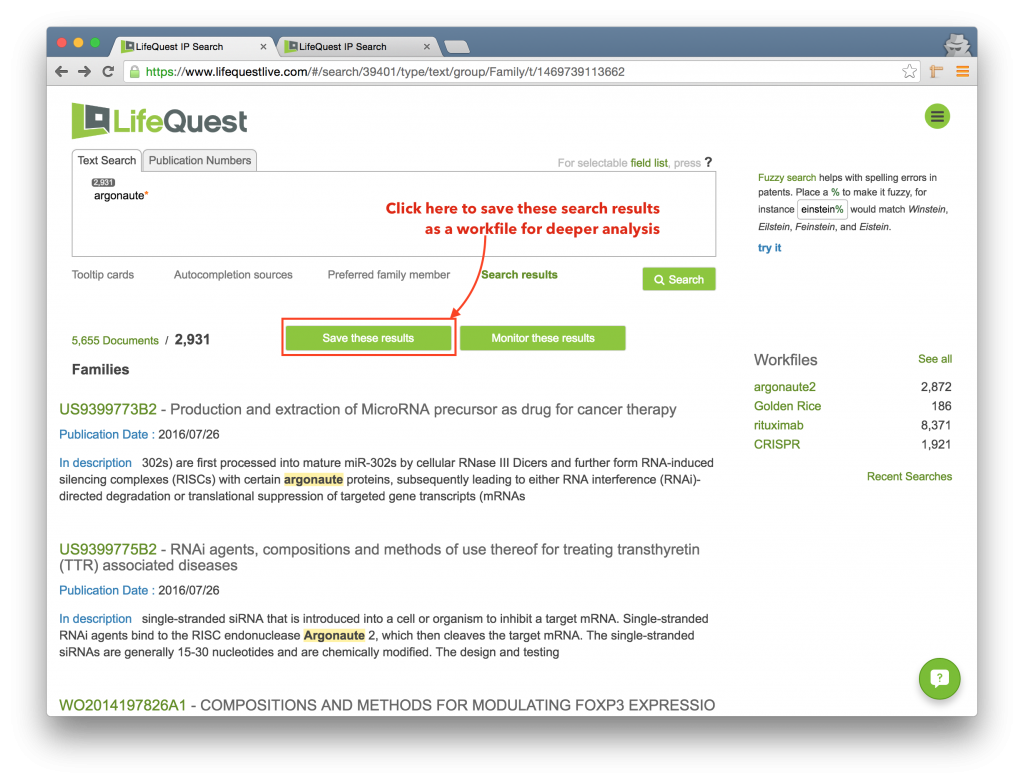
This will pop open a dialog box where you can name your search result.
Once created, you can do deeper analysis on your workfile by clicking its name in the workfile list.